Single-cell transcription group sequencing
Single-cell transcription group sequencing Introduction: 10X GENOMICS is based on The single-cell system of microfluidic technology can simultaneously obtain 100-800,000 cells of the table to achieve the difference in the division of cell populations and the expression of gene expression between cell population, is the heterogeneity of tumor cells. , Immunocytocaproup detection and gold method for embryonic development research. 10xGenomics single-cell applications widely used, applicable cell types: germ cells, embryonic cells, neurons, immunocytes, tumor cells, stem cells, other primary cells. 1
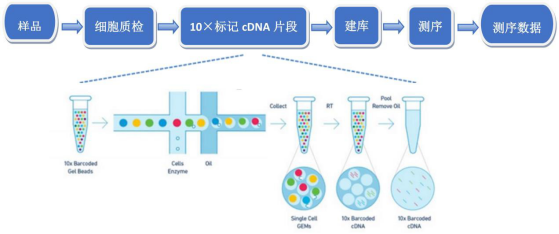
[ 123] 1.1 experimental flow chart [ 1.2 Experiment Description [
1) Cellular Qualification RNA, with COUNTESS u0026 REG; II Automated Cell Counter, the cell concentration was adjusted to the ideal concentration 1 u0026 Times; 106 / ml.2) 10X marked cDNA fragment, gel bead containing Barcode information first combined with mixture of cells and enzymes, and then is located in microfluidic u0026 # 39 ; Double Cross u0026 # 39; Connecting Oil Turne Active Droplets. The droplets are very small, only a gel beads, and the liquid droplets are collected in the reservoir, and the gel beads dissolve the release primer sequence, reverse transcribe the fragment of the cDNA, and perform PCR amplification with cDNA as a template. The product is mixed with Barcode information in each droplet to construct a sequencing literary library. 3) Building a library, first breaking the cDNA into a fragment of from 200 to 300 bp using a Biorupter ultrasonic crusher, plus sequencing joint P5 and sequencing primer R1 After the traditional second-generation sequencing process, the PCR amplification was performed to obtain a DNA library.